Prediction of Patient-Specific Response to Drugs in High Grade Glioma
This site has been active for almost two years. Since that day there have been more than 60,000 different visitors and this month for the first time we will exceed 8,000 visitors. In these two years and despite the pandemic there have been several advances in research even if the impact on clinical practice is slower than what people currently suffering from glioblastoma would expect.
Recently the WHO (World Health Organization) has issued a new classification of high-grade gliomas that distinguishes between mutated IDH glioblastoma and Wild-Type glioblastoma as the prognosis in different cases is very different.
Several times we have mentioned the importance of a personalized medicine to maximize the effectiveness of drugs while reducing the toxicity of treatments. Despite biomarker validation to classify response to treatment, most newly diagnosed (ND) patients receive the same treatment regimen. A recent study sought to determine whether a prospective functional analysis that provided a prediction of the direct drug response based on the specific patient’s tumor cells could accurately predict the clinical response to the drug before treatment.
The study validated a cell culture-based assay for baseline parameters including drug concentrations, timing and reproducibility. Live tumor tissue from patients with high-grade glioma (such as glioblastoma and anaplastic astrocytoma) was tested to establish response parameters. The clinical response between prospective ex vivo response and clinical response was confirmed in newly diagnosed patients enrolled in the 3D-PREDICT clinical trial (NCT03567).
The test accurately predicted response or clinical response to temozolomide in 17/20 (85%) of patients with ND within 7 days of surgery and prior to initiation of treatment. Test responders achieved a median post-intervention overall survival of twice the test predicted non-responders. The case studies provided examples of the clinical utility of dosing predictions and their impact on treatment decisions and led to positive clinical outcomes.
The study was attended by 33 newly diagnosed patients. The following figure illustrates the test results on different drugs as well as on temozolomide (Carbo = carboplatin, Iri = irinotecan, Lom = lomustine, Eto = etoposide, Ever = everolimus, Ruc = rucaparib, Pro = procarbazine, Abe = abemaciclib, Osi = osimertinib, Tra = trametinib). The empty squares indicate the drug has not been tested, the green ones the responders (according to the test), the yellow ones the moderate responders and finally the red ones the non-responders. The rows indicate the specific cases and for each of these the histology (AA = Anaplastic Astrocytoma or GBM = glioblastoma), the specific mutations (IDH = IDH mutated or WT = wild type) and the mediation status (U = not methylated, M = methylated).
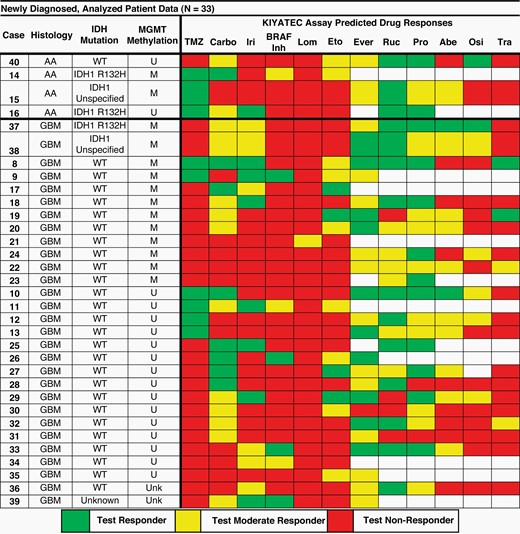
The results were so compelling that KIYATEC, the innovative company in functional precision oncology that developed the system, announced that it has made the 3D-PREDICT Glioma test available and accessible to the public.
Sounds like great news to me. I ask you again to continue to help the fundraising campaign Glioblastoma.it for CUSP-ND for Emanuele by sharing the link in order to spread the word and raise awareness as many people as possible.